Mucus Secretion in Lung Health and Disease
Institution: Ulm University
Timeframe: 07/2023 – 07/2024
Keywords: Mucus secretion, SNARE complex, NHBE, ALI culture, ELISA, Western Blot, qPCR, FACS, mechanistic biology, mucociliary clearance
Overview:
This project focused on characterizing basal and stimulated mucus secretion in primary human airway epithelial cells (NHBE) cultured at the air-liquid interface (ALI). I investigated the molecular machinery regulating vesicle fusion, particularly the SNARE complex (including SYT and STX isoforms). The work included mechanistic assays and analysis of human donor variability. This experience sparked my interest in immunosenescence, inflammation-driven aging, and senolytics.
Techniques & Tools:
- ELISA-based mucus quantification
- Western blotting (SNARE complex components)
- qRT-PCR (384-well format)
- Multicolor flow cytometry
- Cell sorting and assay optimization
- Protein concentration assays
- Lab automation using 384-well robotic pipetting systems
- Basic coding (data preprocessing, plate layout design)
Key Learnings:
- Uncovered the complexity of IL-13 batch-to-batch variability in mucus stimulation assays.
- Developed critical insights into the link between chronic inflammation and cellular aging.
- Identified flow cytometry as my central analytical skill.
- Conceptual foundation for my current senescence-targeted CAR-T cell therapy interest.
Optogenetic Regulation of Angiogenesis
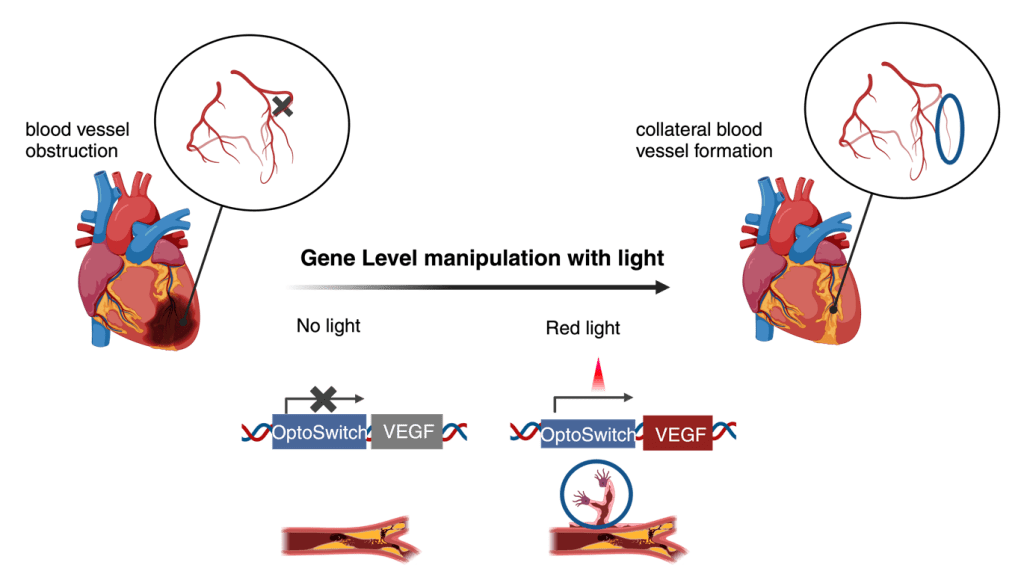
Institution: German Primate Center (DPZ) & Max Planck Institute for Dynamics and Self-Organization
Timeframe: 03/2022 – 09/2022
Keywords: Optogenetics, endothelial cells, rodent model, angiogenesis, red light control, molecular cloning, microscopy
Overview:
For my master’s thesis, I studied red light-controlled angiogenesis in endothelial cells isolated from explanted rodent aortas. The project utilized optogenetic construct (Optoswitch) to manipulate endothelial signaling and migration in response to light stimuli. I compared different transfection strategies across rodent and porcine models, gaining firsthand experience in the challenges of primary cell gene delivery.
Techniques & Tools:
- Molecular cloning and SnapGene construct design
- Brightfield and fluorescence microscopy
- Aortic dissection from rodent models
- Optogenetic stimulation (red light modulation)
- Scratch (wound healing) assays
- Viral vector troubleshooting
- Gene expression analysis via qPCR
- Data visualization using GraphPad Prism
Key Learnings:
- Highlighted the limitations of viral delivery in primary endothelial cultures.
- Deepened my fascination with logic-based optogenetic control of cellular function.
- Understood how culture conditions profoundly affect angiogenesis outcomes.
THP-1 Macrophage Differentiation and Co-Culture (cell culture for another student/Project)
Institution: DPZ
Keywords: THP-1, macrophage polarization, cytokine stimulation, epithelial–immune interaction
Overview:
Because of kindness of postdoc in our depratment, I practiced cell culture of THP-1 macrophage model to study immune-endothelial l cross-talk in cardiovascular inflammation. Differentiation was induced via PMA followed by polarization using IFN-γ or IL-4/IL-13. The model was applied to mimic blood vessel inflammation.
Techniques & Tools:
- THP-1 differentiation protocols
- Cytokine stimulation (M1/M2 polarization)
- Co-culturing with endothelial cells
Key Learnings:
- Built a strong foundation in innate immune modeling.
- Enhanced my co-culture assay design skills.
Gene Transfer and Live-Cell Monitoring
Institution: Various labs during Master’s program
Keywords: Viral transduction, CRISPR, cardiac inflammation, live imaging, STAT signaling
Overview:
Across three lab rotations, I assisted in gene transfer and inflammatory pathway studies. One focus was CRISPR editing using AAV vectors, another targeting m6A methylation regulators in rodent cardiac tissue. Third focused on STAT signaling in inflammation.
Techniques & Tools:
- AAV-mediated gene transfer
- CRISPR editing
- Live-cell imaging
- Cell viability assays
Key Learnings:
- Recognized the translational limitations of current gene delivery methods.
- Reinforced my goal to engineer controllable, modular cell therapies.
📌 Skills Summary (Built from Past Projects)
- Flow Cytometry & Cell Sorting:
MACSQuant 10, Navios EX, FACS Symphony | Multicolor panels | Surface/intracellular staining | Cell sorting - Molecular & Protein Techniques:
qRT-PCR (including 384-well automation) | Western blotting | ELISA | Cytokine profiling - Cell Culture & Co-Culture Models:
NHBE ALI cultures | THP-1 macrophage models | Primary endothelial and epithelial cells | Transwell assays - Gene & Protein Delivery:
Viral vector transduction | CRISPR/AAV | Optogenetics - Microscopy & Imaging:
Live-cell imaging | Fluorescence & brightfield microscopy | Angiogenesis and wound healing assays - Automation & Data Analysis:
Robotic pipetting (384-well) | LIMS | GraphPad Prism | Basic coding (Rstudio/Bioconductor, Excel macros)